[new preprint] Fast reconstruction of degenerate populations of conductance-based neuron models from spike times
My first preprint is now available on arXiv! with Damien Ernst, Guillaume Drion, and Arthur Fyon.
I am excited to share my FIRST preprint that is now available on arXiv ! You can also find it on Orbi with the supplementary materials: ORBi.
Author summary
Neurons communicate through spikes, and spike timing is a crucial part of neuronal processing. Spike times can be recorded experimentally both intracellularly and extracellularly, and are the main output of state-of-the-art neural probes. On the other hand, neuronal activity is controlled at the molecular level by the currents generated by many different transmembrane proteins called ion channels. Connecting spike timing to ion channel composition remains an arduous task to date. To address this challenge, we developed a method that combines deep learning with a theoretical tool called Dynamic Input Conductances (DICs), which reduce the complexity of ion channel interactions into three interpretable components describing how neurons spike. Our approach uses deep learning to infer DICs directly from spike times and then generates populations of “twin” neuron models that replicate the observed activity while capturing natural variability in membrane channel composition. The method is fast, accurate, and works using only spike recordings. We also provide open-source software with a graphical interface, making it accessible to researchers without programming expertise.
Abstract
Inferring the biophysical parameters of conductance-based models (CBMs) from experimentally accessible recordings remains a central challenge in computational neuroscience. Spike times are the most widely available data, yet they reveal little about which combinations of ion channel conductances generate the observed activity. This inverse problem is further complicated by neuronal degeneracy, where multiple distinct conductance sets yield similar spiking patterns. We introduce a method that addresses this challenge by combining deep learning with Dynamic Input Conductances (DICs), a theoretical framework that reduces complex CBMs to three interpretable feedback components governing excitability and firing patterns. Our approach first maps spike times directly to DIC values at threshold using a lightweight neural network that learns a low-dimensional representation of neuronal activity. The predicted DIC values are then used to generate degenerate CBM populations via an iterative compensation algorithm, ensuring compatibility with the intermediate target DICs, and thereby reproducing the corresponding firing patterns, even in high-dimensional models. Applied to two neuronal models, this algorithmic pipeline reconstructs spiking, bursting, and irregular regimes with high accuracy and robustness to variability, including spike trains generated by Poisson processes. It produces diverse degenerate populations within milliseconds on standard hardware, enabling scalable and efficient inference from spike recordings alone. Beyond methodological advances, we provide an open-source software package with a graphical interface that allows experimentalists to generate and explore CBM populations directly from spike trains without requiring programming expertise. Together, this work positions DICs as a practical and interpretable link between experimentally observed activity and mechanistic models. By enabling fast and scalable reconstruction of degenerate populations directly from spike times, our approach provides a powerful way to investigate how neurons exploit conductance variability to achieve reliable computation and provides the foundation for experimental applications that span from neuromodulation studies to real-time model-guided interventions.
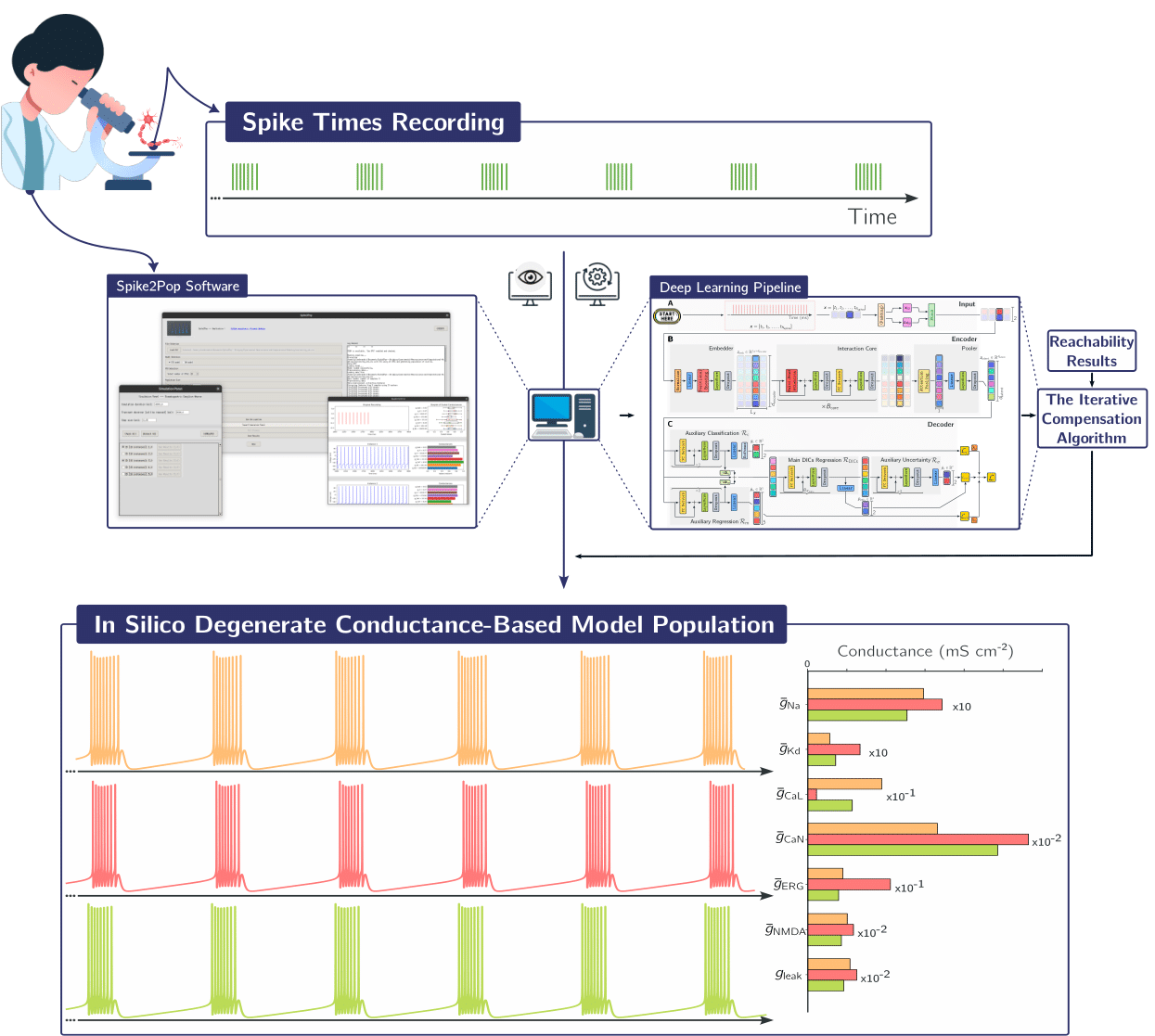 Figure 1: Our approach is designed to be practical for experimentalists through a user-friendly interface. Starting from spike train recordings, users can easily generate and explore populations of conductance-based models (CBMs) that replicate the observed spiking activity.
Figure 1: Our approach is designed to be practical for experimentalists through a user-friendly interface. Starting from spike train recordings, users can easily generate and explore populations of conductance-based models (CBMs) that replicate the observed spiking activity.
Contact: For questions or collaborations, please reach out to me at jbrandoit@uliege.be.
Damien Ernst’s webpage : Damien Ernst
Guillaume Drion’s webpage : Guillaume Drion
Arthur Fyon’s webpage : Arthur Fyon
BibTeX citation:
@misc{brandoit2025fastreconstructiondegeneratepopulations,
title={Fast reconstruction of degenerate populations of conductance-based neuron models from spike times},
author={Julien Brandoit and Damien Ernst and Guillaume Drion and Arthur Fyon},
year={2025},
eprint={2509.12783},
archivePrefix={arXiv},
primaryClass={q-bio.NC},
url={https://arxiv.org/abs/2509.12783},
}